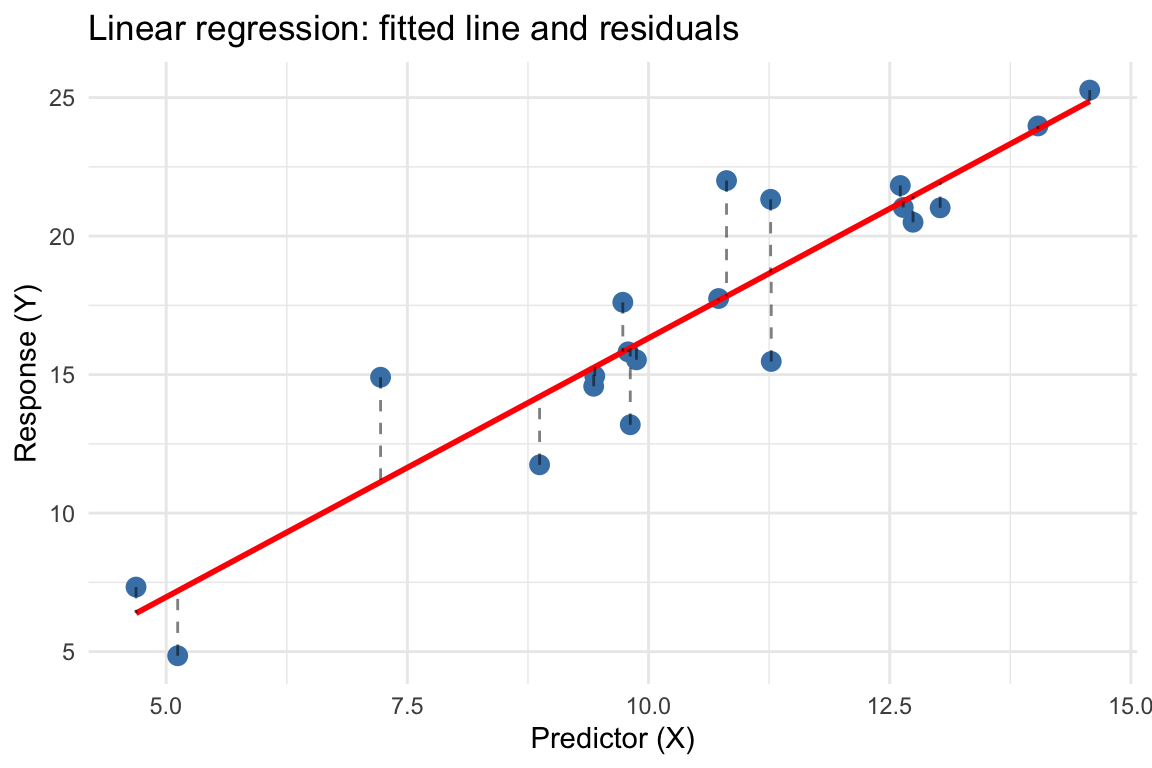
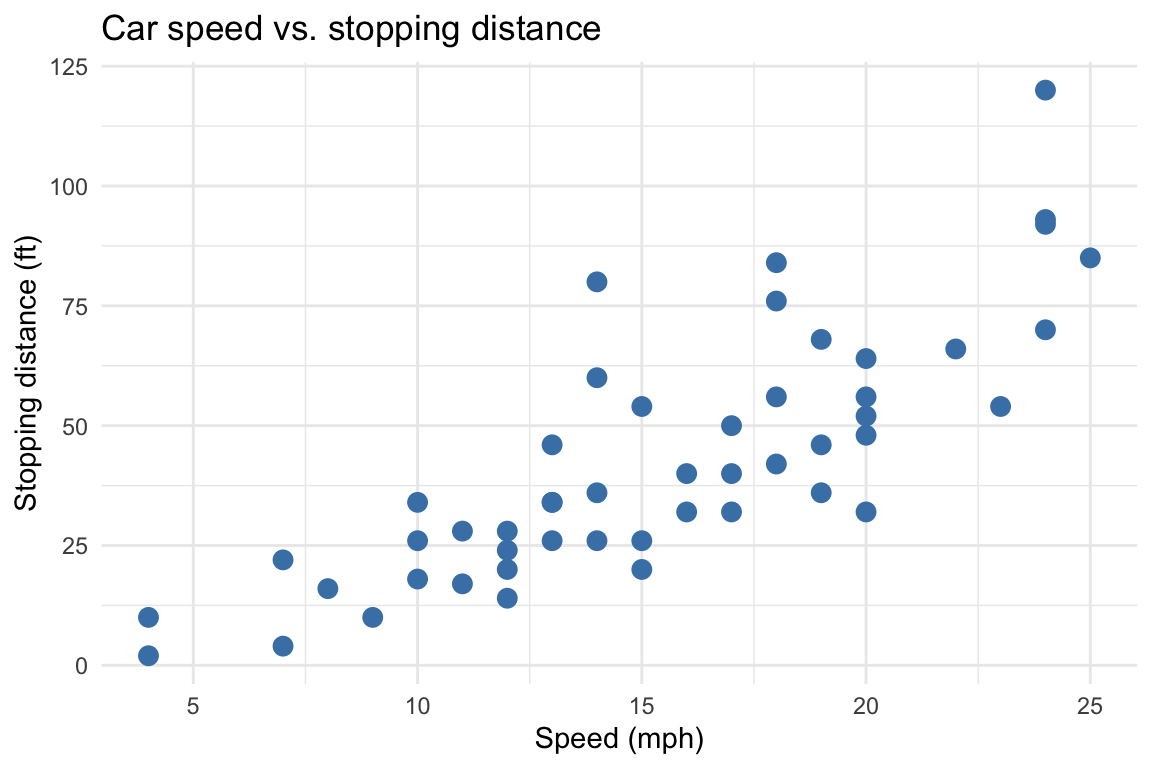
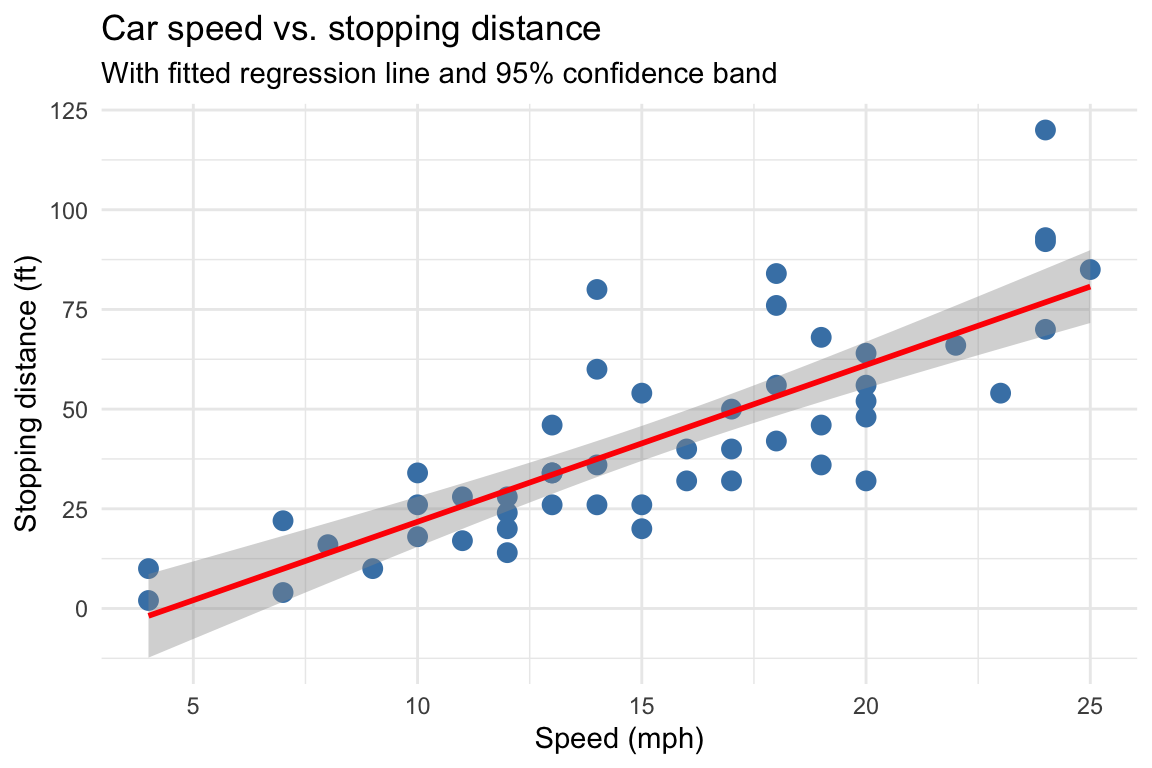
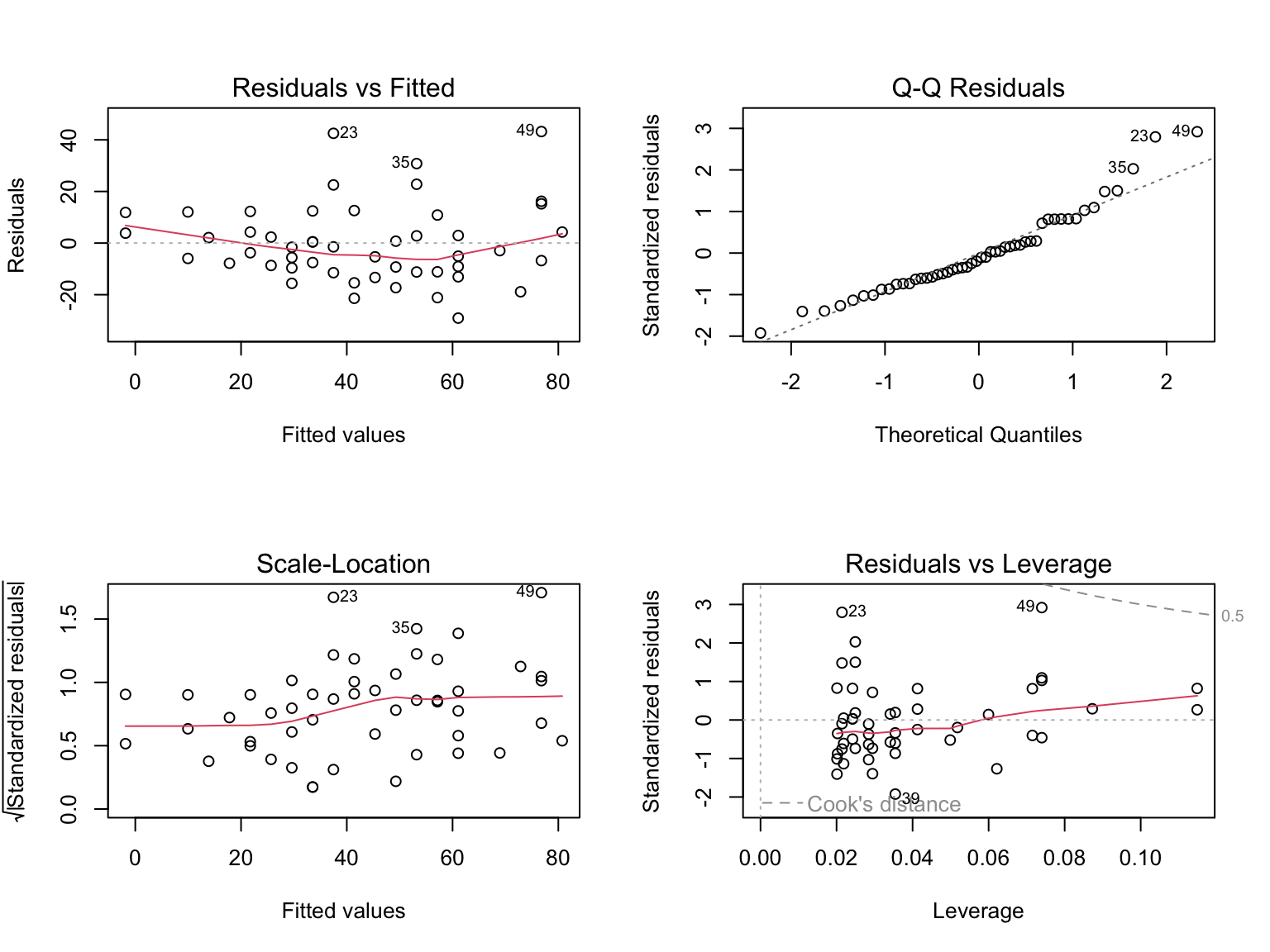
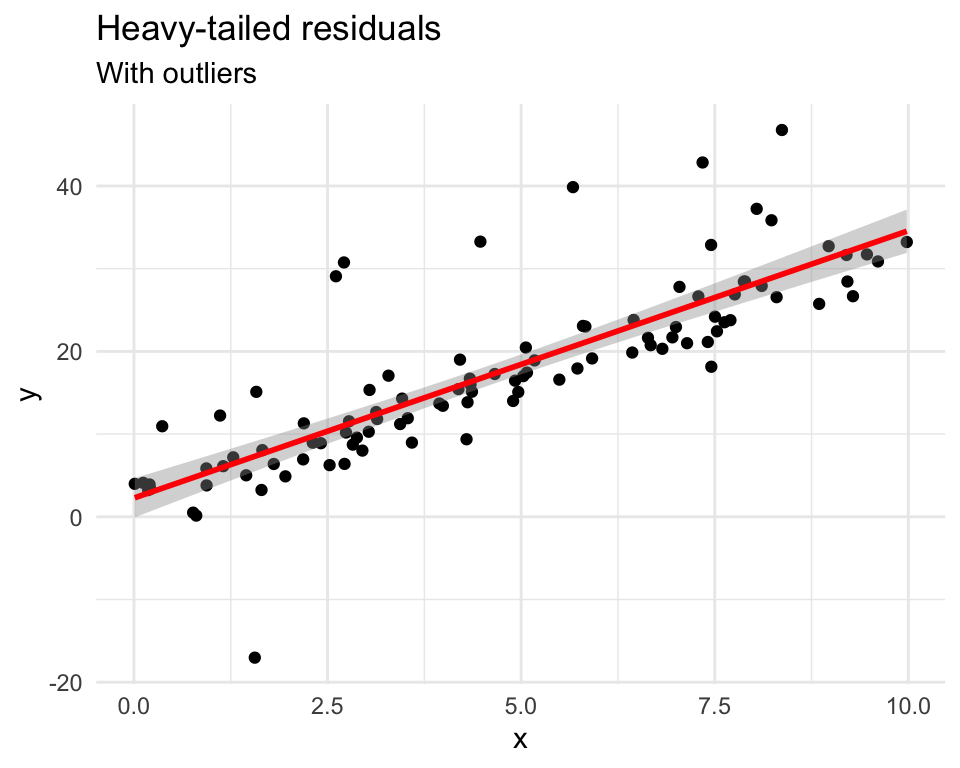
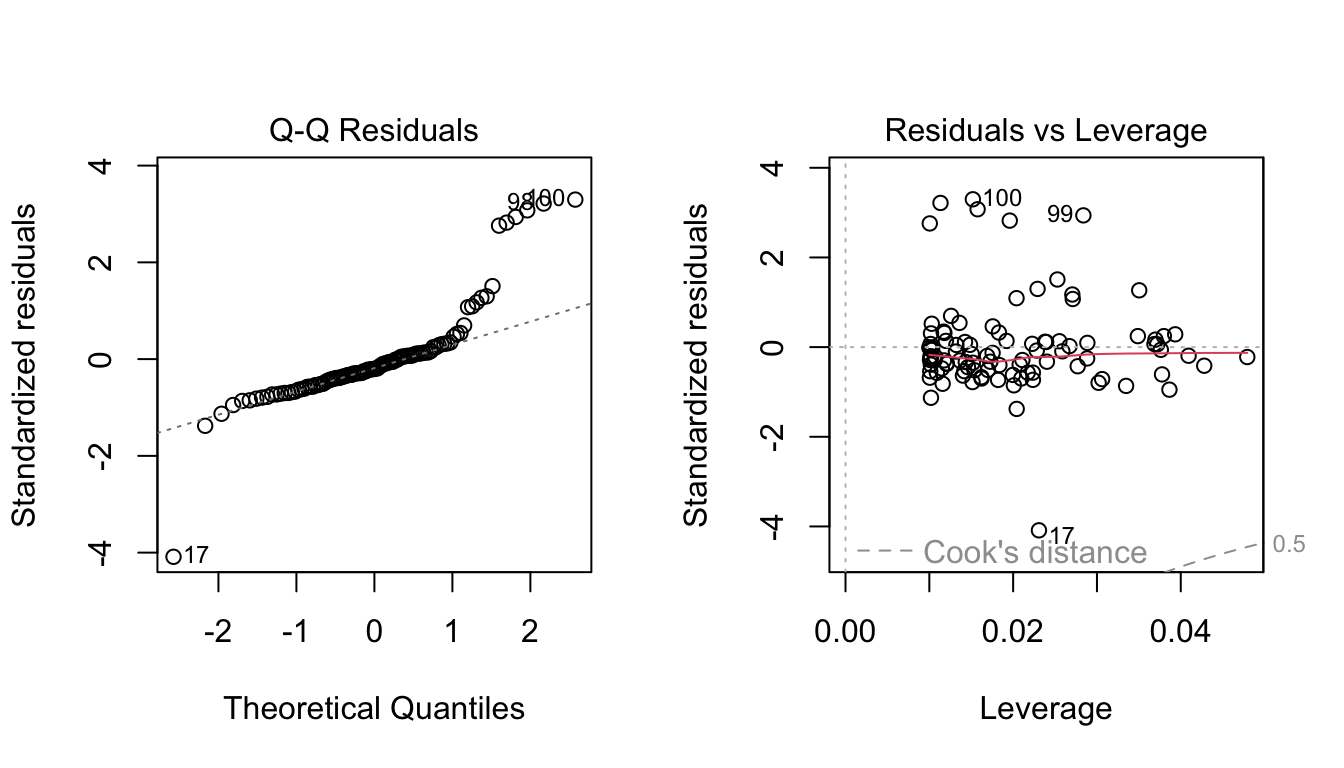
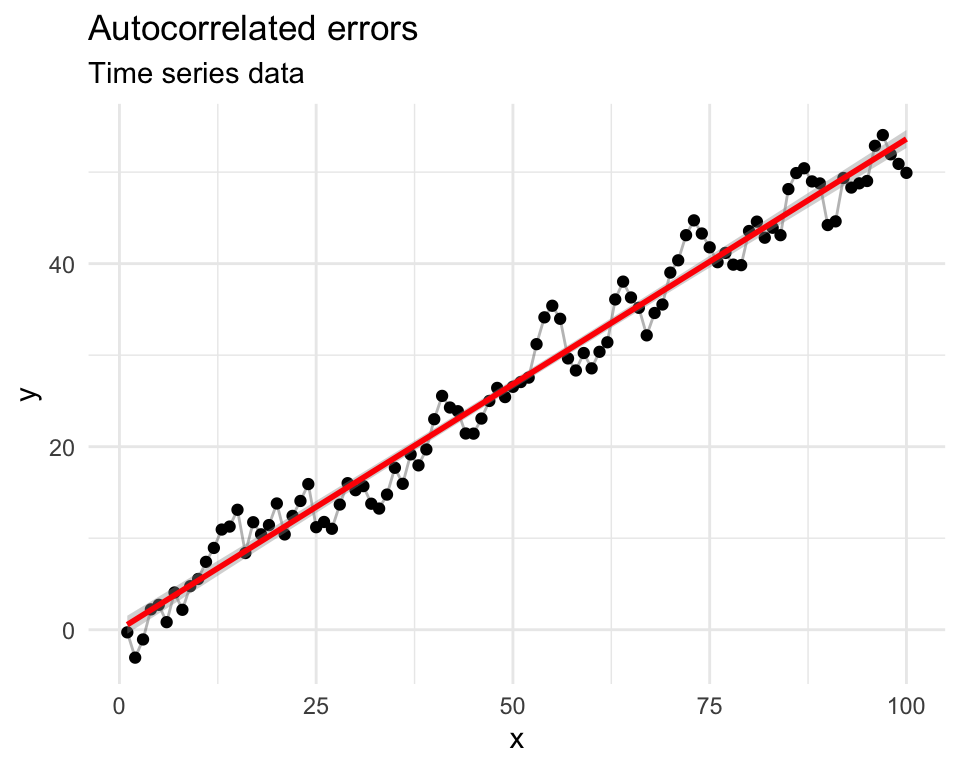
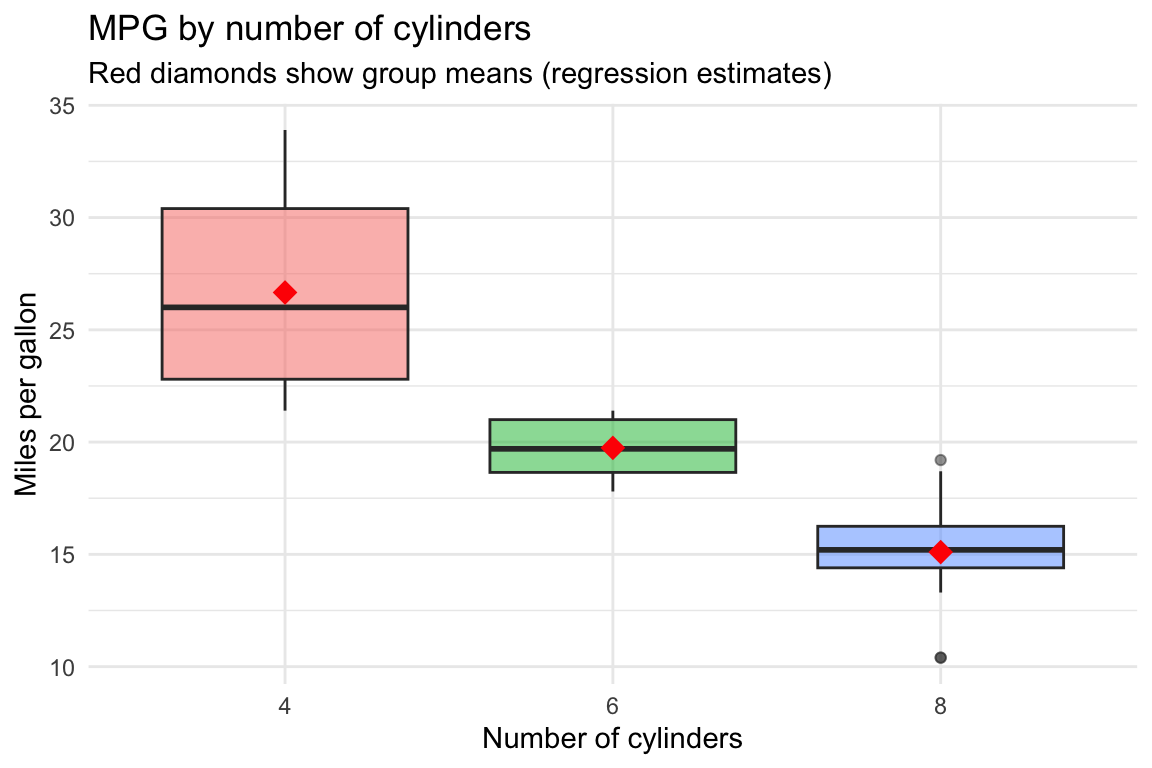
set.seed(42)
x <- rnorm(20, mean = 10, sd = 2)
y <- 2 + 1.5 * x + rnorm(20, mean = 0, sd = 2)
df <- data.frame(x = x, y = y)
fit <- lm(y ~ x, data = df)
ggplot(df, aes(x = x, y = y)) +
geom_point(size = 3, color = "steelblue") +
geom_smooth(method = "lm", se = FALSE, color = "red", linewidth = 1) +
geom_segment(aes(xend = x, yend = fitted(fit)),
linetype = "dashed", alpha = 0.5) +
labs(title = "Linear regression: fitted line and residuals",
x = "Predictor (X)", y = "Response (Y)") +
theme_minimal()