Call:
glm(formula = cbind(num_toxicities, num_patients - num_toxicities) ~
I(log(dose)), family = binomial(), data = dlt_data)
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -3.3528 1.6537 -2.027 0.0426 *
I(log(dose)) 0.9309 0.6495 1.433 0.1518
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 10.159 on 5 degrees of freedom
Residual deviance: 7.476 on 4 degrees of freedom
AIC: 16.751
Number of Fisher Scoring iterations: 5Week 13: Logistic Regression & AI-assisted Workflows
Part 1: Logistic regression
Overview
- Recall from Week 9: in linear regression, we modeled a continuous response \(Y\) as \[Y = \beta_0 + \beta_1 X + \epsilon, \] where \(\epsilon\) was a normally distributed error term. Estimation and inference used least squares and normal-error assumptions.
New question
What if the response \(Y\) is binary (0/1)?
- The “mean” of \(Y\) is now a probability that \(Y = 1\)
- Residuals cannot possibly be normal in this situation
- Predictions for the mean could fall outside \((0, 1)\)
Better model: logistic regression…
Motivating example: Dose–toxicity modeling
We consider an early-phase Oncology clinical trial where cohorts of patients receive increasing drug doses. The binary outcome \(Y\) indicates whether a dose-limiting toxicity (DLT) occurred (1 = yes, 0 = no).
Construct dose-toxicity dataset (dlt_data) and display empirical rates.
dlt_data <- tibble::tibble(
dose = c(1, 2.5, 5, 10, 20, 25),
num_patients = c(3, 4, 5, 4, 6, 2),
num_toxicities = c(0, 1, 0, 1, 1, 2)
) |>
dplyr::mutate(empirical_rate = num_toxicities / num_patients)
dlt_data |>
gt() |>
tab_header(title = "Drug A Dose–Toxicity Data (DLTs)") |>
cols_label(
dose = "Dose A (mg)",
num_patients = "Patients",
num_toxicities = "DLTs",
empirical_rate = "Proportion of DLTs"
) |>
fmt_percent(columns = empirical_rate, decimals = 1) |>
fmt_number(columns = c(num_patients, num_toxicities), decimals = 0) |>
tab_style(
style = cell_fill(color = "#f2f7ff"),
locations = cells_column_labels()
) |>
opt_table_font(font = "Arial")| Drug A Dose–Toxicity Data (DLTs) | |||
| Dose A (mg) | Patients | DLTs | Proportion of DLTs |
|---|---|---|---|
| 1.0 | 3 | 0 | 0.0% |
| 2.5 | 4 | 1 | 25.0% |
| 5.0 | 5 | 0 | 0.0% |
| 10.0 | 4 | 1 | 25.0% |
| 20.0 | 6 | 1 | 16.7% |
| 25.0 | 2 | 2 | 100.0% |
Fitting logistic regression in R
Making predictions
Plotting the predictions
Plot on probability scale
# Probability scale
dose_grid <- tibble::tibble(dose = seq(min(dlt_data$dose), max(dlt_data$dose), length.out = 100)) |>
dplyr::mutate(p_hat = predict(fit_dose, newdata = dplyr::cur_data(), type = "response"))
ggplot(dlt_data, aes(x = dose, y = empirical_rate)) +
geom_point(size = 3, color = "#1f77b4") +
geom_line(data = dose_grid, aes(y = p_hat), color = "#d62728", linewidth = 1) +
scale_y_continuous(labels = scales::percent_format(accuracy = 1)) +
scale_x_log10() +
labs(title = "Probability scale",
x = "Dose A (mg)",
y = "Probability of DLT") +
theme_minimal()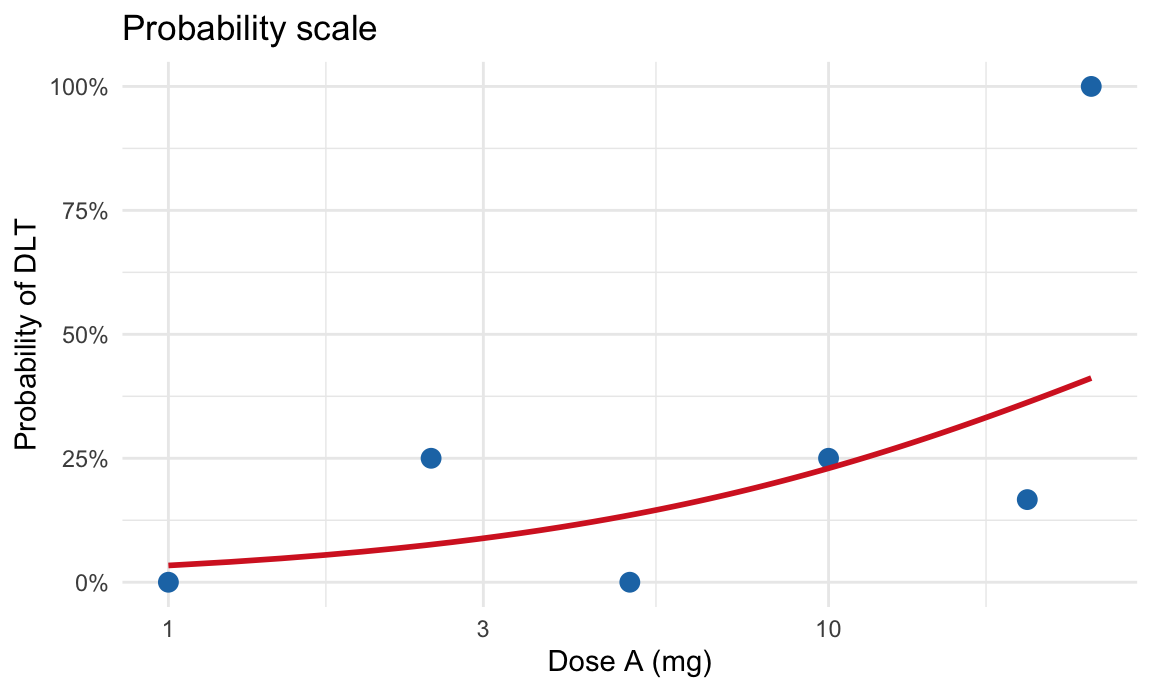
Plot on linear predictor scale
# Linear predictor (log-odds) scale
dose_grid_lp <- tibble::tibble(dose = seq(min(dlt_data$dose), max(dlt_data$dose), length.out = 100))
pred_lp <- predict(fit_dose, newdata = dose_grid_lp, type = "link", se.fit = TRUE)
dose_grid_lp$eta_hat <- as.numeric(pred_lp$fit)
ggplot(dose_grid_lp, aes(x = dose, y = eta_hat)) +
geom_line(color = "#d62728", linewidth = 1) +
scale_x_log10() +
labs(title = "Linear predictor scale",
x = "Dose A (mg)",
y = "Log-odds of DLT") +
theme_minimal()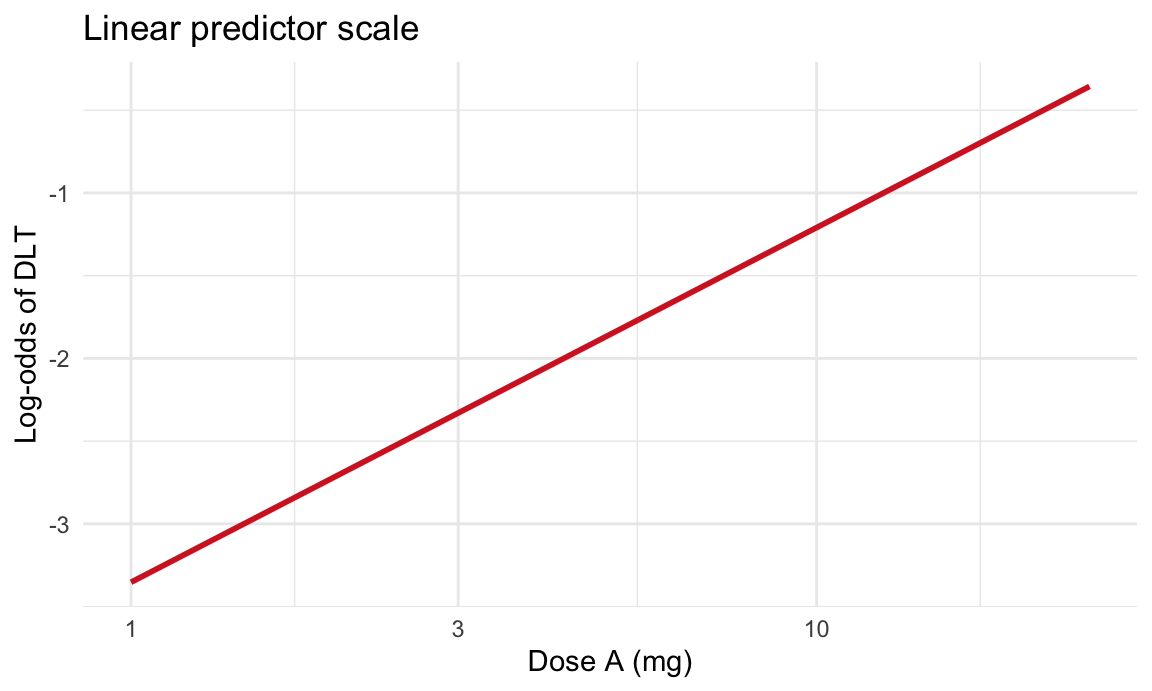
Generalized linear models (GLMs)
- A unified framework that includes linear, logistic, and Poisson regression (among others).
| Model | Typical data | R syntax | Link function |
|---|---|---|---|
| Linear regression | Continuous outcomes (approximately normal residuals) | lm(y ~ x, data = df)glm(y ~ x, data = df, family = gaussian()) |
identity |
| Logistic regression | Binary 0/1 outcomes or successes/trials | glm(y ~ x, family = binomial(), data = df)glm(cbind(successes, trials - successes) ~ x, family = binomial(), data = df) |
logit |
| Poisson regression | Count data | glm(y ~ x, family = poisson(), data = df) |
log |
Part 2: AI-assisted workflows with R
Where do humans1 need AI?
- Learning with AI
- Code generation
- Documentation and testing of code
Where does AI need humans?
- Trust
- Integration
- Taste
Trust
- How do your stakeholders know a statistical result is trustworthy?
- Who is accountable for correctness? Must be a human
- Must be able to identify issues in e.g. statistical ideas and/or generated code
Integration
- Effective/impactful use of AI requires a new class of experts, e.g.
- Technical experts: technology underlying LLMs is itself statistical (models trained on massive data, predicting the next word)
- Facilitators: people who understand what tools are available and can connect them to solve practical problems
Taste
- Knowing what to ask (prompt engineering)
- Discriminating between good/useful output, and poor/unsuitable output
- Knowing when AI should not be used
What does it mean for me and you?
- Need knowledge and experience to develop taste, e.g.
- Statistical knowledge
- R programming experience
- Need to learn how and when to effectively work with AI