Week 10: Regression (continued)
Recall: What is regression about?
- Modeling the relationship between variables
- Response variable (dependent variable, outcome): what we want to predict or explain
- Predictor variable(s) (independent variable(s), explanatory variable(s)): what we use to predict or explain
- Potential goals:
- Prediction: estimate the response for new observations
- Inference: understand whether and how predictors affect the response
Multiple regression
Extending to multiple predictors
Multiple regression includes more than one predictor \[Y = \beta_0 + \beta_1 X_1 + \beta_2 X_2 + \cdots + \beta_p X_p + \epsilon\]
Each coefficient \(\beta_j\) represents the effect of \(X_j\) on \(Y\), holding other predictors constant
R syntax:
Example: mtcars dataset
mpg cyl disp hp drat wt qsec vs am gear carb
Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4
Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4
Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1
Hornet 4 Drive 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1
Hornet Sportabout 18.7 8 360 175 3.15 3.440 17.02 0 0 3 2
Valiant 18.1 6 225 105 2.76 3.460 20.22 1 0 3 1mpg: miles per gallonwt: weight (1000 lbs)hp: horsepowercyl: number of cylinders
Research question
- How do weight and horsepower jointly affect fuel efficiency (mpg)? \[\text{mpg} = \beta_0 + \beta_1 \times \text{wt} + \beta_2 \times \text{hp} + \epsilon\]
Fitting the multiple regression model
Call:
lm(formula = mpg ~ wt + hp, data = mtcars)
Residuals:
Min 1Q Median 3Q Max
-3.941 -1.600 -0.182 1.050 5.854
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 37.22727 1.59879 23.285 < 2e-16 ***
wt -3.87783 0.63273 -6.129 1.12e-06 ***
hp -0.03177 0.00903 -3.519 0.00145 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 2.593 on 29 degrees of freedom
Multiple R-squared: 0.8268, Adjusted R-squared: 0.8148
F-statistic: 69.21 on 2 and 29 DF, p-value: 9.109e-12Interpreting multiple regression coefficients
- Intercept (\(\hat{\beta}_0 = 37.23\)): expected mpg when weight and horsepower are both 0 (not meaningful)
- wt (\(\hat{\beta}_1 = -3.88\)): for each 1000 lb increase in weight, mpg decreases by about 3.88, holding horsepower constant
- hp (\(\hat{\beta}_2 = -0.03\)): for each 1 hp increase, mpg decreases by about 0.03, holding weight constant
Comparing models
- We can compare models with different numbers of predictors
[1] 0.7528328[1] 0.8267855- Model 2 explains more variance (higher R-squared)
ANOVA for model comparison
Analysis of Variance Table
Model 1: mpg ~ wt
Model 2: mpg ~ wt + hp
Res.Df RSS Df Sum of Sq F Pr(>F)
1 30 278.32
2 29 195.05 1 83.274 12.381 0.001451 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1- The ANOVA F-test compares the two models
- Small p-value indicates that adding
hpsignificantly improves the model - The more complex model (model2) is preferred
Categorical predictors
Including categorical variables
- Categorical predictors are automatically converted to dummy variables
- Example: modeling mpg as a function of number of cylinders
cylcan be 4, 6, or 8
Call:
lm(formula = mpg ~ cyl, data = mtcars)
Residuals:
Min 1Q Median 3Q Max
-5.2636 -1.8357 0.0286 1.3893 7.2364
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 26.6636 0.9718 27.437 < 2e-16 ***
cyl6 -6.9208 1.5583 -4.441 0.000119 ***
cyl8 -11.5636 1.2986 -8.905 8.57e-10 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 3.223 on 29 degrees of freedom
Multiple R-squared: 0.7325, Adjusted R-squared: 0.714
F-statistic: 39.7 on 2 and 29 DF, p-value: 4.979e-09Interpreting categorical coefficients
- R creates dummy variables with the first level as the reference category
- Here, 4 cylinders is the reference
- Intercept (\(\hat{\beta}_0 = 26.66\)): mean mpg for 4-cylinder cars
- cyl6 (\(\hat{\beta}_1 = -6.92\)): 6-cylinder cars have about 6.92 lower mpg than 4-cylinder cars, on average
- cyl8 (\(\hat{\beta}_2 = -11.56\)): 8-cylinder cars have about 11.56 lower mpg than 4-cylinder cars, on average
Visualizing categorical regression
Boxplot with group means
ggplot(mtcars, aes(x = cyl, y = mpg, fill = cyl)) +
geom_boxplot(alpha = 0.5) +
stat_summary(fun = mean, geom = "point", shape = 18, size = 4, color = "red") +
labs(title = "MPG by number of cylinders",
subtitle = "Red diamonds show group means (regression estimates)",
x = "Number of cylinders", y = "Miles per gallon") +
theme_minimal() +
theme(legend.position = "none")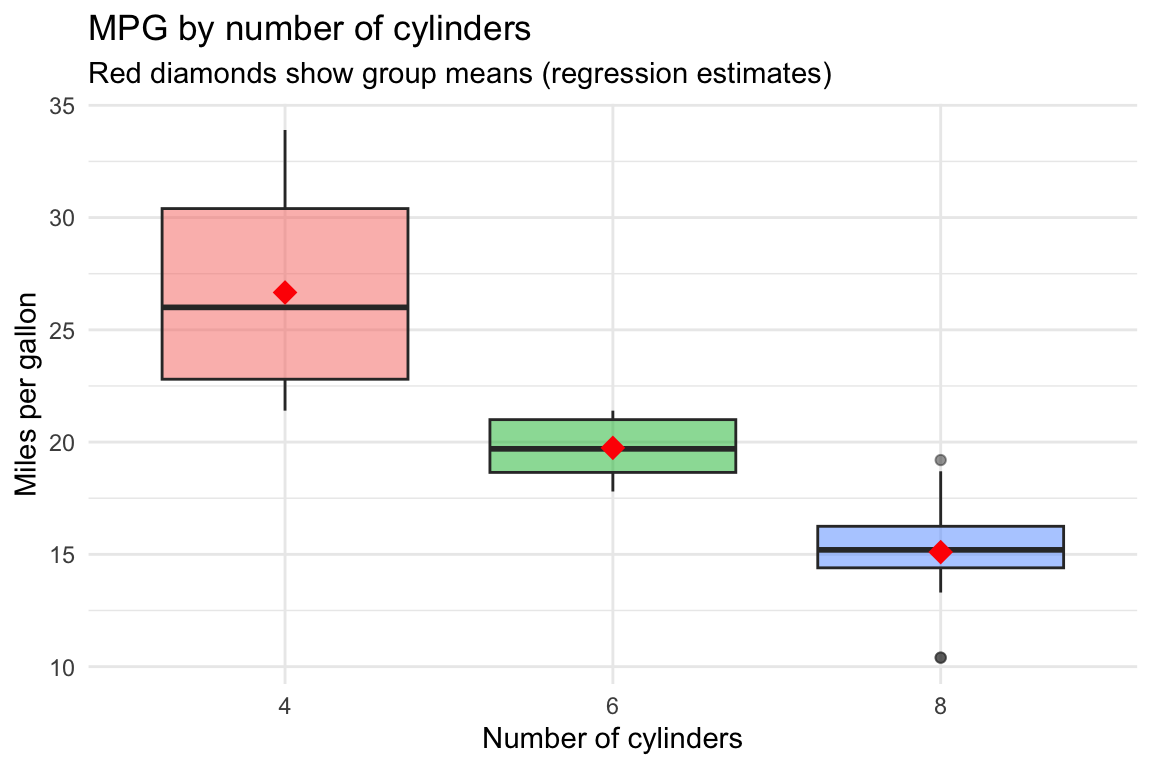
Key R functions for regression
Summary table
| Function | Purpose | Example |
|---|---|---|
lm() |
Fit linear model | lm(y ~ x, data = df) |
summary() |
Model summary statistics | summary(model) |
coef() |
Extract coefficients | coef(model) |
confint() |
Confidence intervals | confint(model) |
predict() |
Make predictions | predict(model, newdata) |
plot() |
Diagnostic plots | plot(model) |
anova() |
Compare models | anova(model1, model2) |
Formula syntax in R
| Formula | Meaning |
|---|---|
y ~ x |
Simple regression: y on x |
y ~ x1 + x2 |
Multiple regression: y on x1 and x2 |
y ~ x1 + x2 + x1:x2 |
Include interaction between x1 and x2 |
y ~ x1 * x2 |
Equivalent to y ~ x1 + x2 + x1:x2 |
y ~ . |
Use all other variables as predictors |
y ~ x - 1 |
Regression without intercept |
Important considerations
Correlation vs. causation
- Regression reveals associations, not necessarily causation
- Just because \(X\) predicts \(Y\) doesn’t mean \(X\) causes \(Y\)
- Possible explanations for correlation:
- \(X\) causes \(Y\)
- \(Y\) causes \(X\)
- A third variable causes both \(X\) and \(Y\) (confounding)
- The association is due to chance
- Randomized experiments (like the clinical trials in Week 4) provide stronger evidence for causation
Extrapolation
- Extrapolation: making predictions outside the range of observed data
- The linear relationship may not hold beyond the observed range
- Example: predicting stopping distance for a car going 100 mph using our cars model
- The data only include speeds from 4 to 25 mph
- Extrapolating to 100 mph is risky
Limitations of R-squared
- R-squared measures the proportion of variance explained
- Higher R-squared doesn’t always mean a better model:
- Adding more predictors always increases R-squared
- A model with lower R-squared might generalize better
- R-squared doesn’t tell us if the model assumptions are met
- Use adjusted R-squared when comparing models with different numbers of predictors
- Penalizes for adding unnecessary predictors
Example: Full analysis
Palmer Penguins dataset
Load and display the penguins dataset
tibble [344 × 8] (S3: tbl_df/tbl/data.frame)
$ species : Factor w/ 3 levels "Adelie","Chinstrap",..: 1 1 1 1 1 1 1 1 1 1 ...
$ island : Factor w/ 3 levels "Biscoe","Dream",..: 3 3 3 3 3 3 3 3 3 3 ...
$ bill_length_mm : num [1:344] 39.1 39.5 40.3 NA 36.7 39.3 38.9 39.2 34.1 42 ...
$ bill_depth_mm : num [1:344] 18.7 17.4 18 NA 19.3 20.6 17.8 19.6 18.1 20.2 ...
$ flipper_length_mm: int [1:344] 181 186 195 NA 193 190 181 195 193 190 ...
$ body_mass_g : int [1:344] 3750 3800 3250 NA 3450 3650 3625 4675 3475 4250 ...
$ sex : Factor w/ 2 levels "female","male": 2 1 1 NA 1 2 1 2 NA NA ...
$ year : int [1:344] 2007 2007 2007 2007 2007 2007 2007 2007 2007 2007 ...Research question
- How does body mass depend on flipper length and species?
- Let’s build a multiple regression model
Exploratory visualization
Scatterplot colored by species
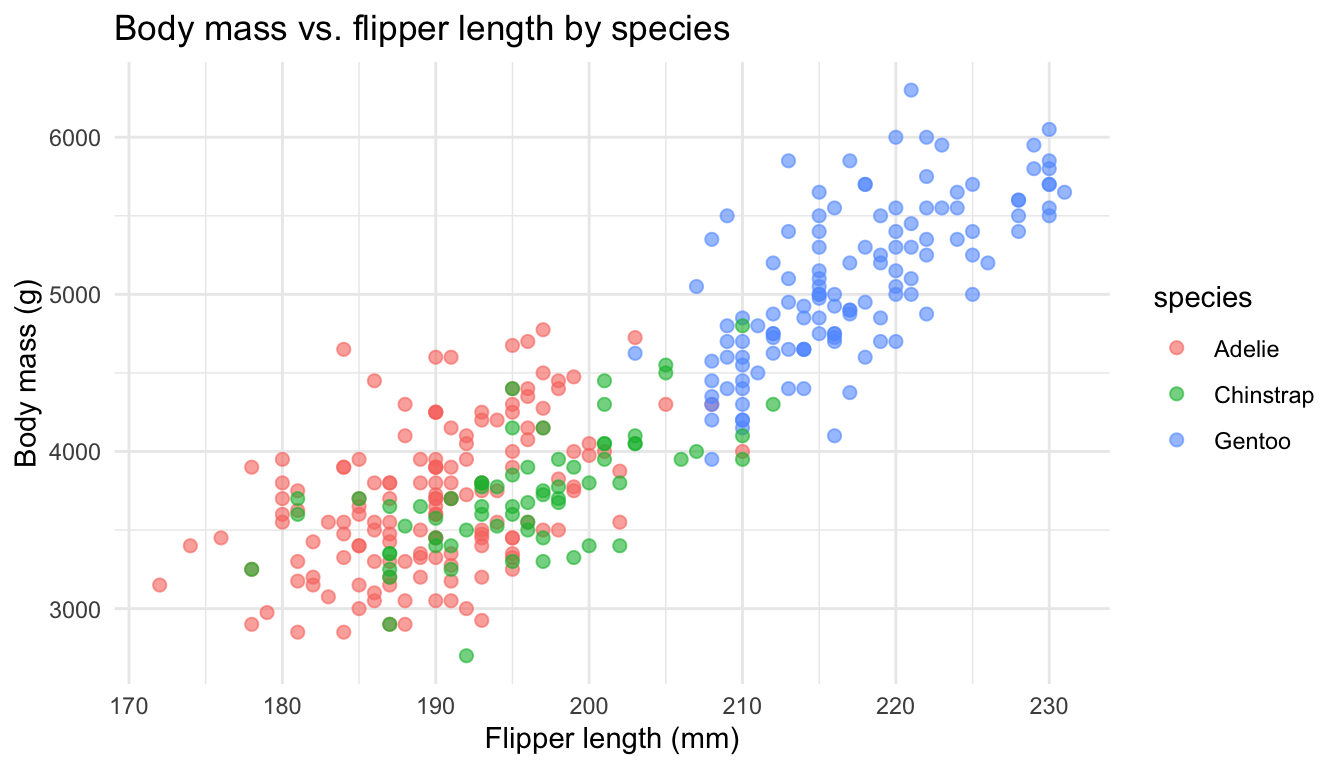
Fitting the model
Call:
lm(formula = body_mass_g ~ flipper_length_mm + species, data = penguins_clean)
Residuals:
Min 1Q Median 3Q Max
-927.70 -254.82 -23.92 241.16 1191.68
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -4031.477 584.151 -6.901 2.55e-11 ***
flipper_length_mm 40.705 3.071 13.255 < 2e-16 ***
speciesChinstrap -206.510 57.731 -3.577 0.000398 ***
speciesGentoo 266.810 95.264 2.801 0.005392 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 375.5 on 338 degrees of freedom
Multiple R-squared: 0.7826, Adjusted R-squared: 0.7807
F-statistic: 405.7 on 3 and 338 DF, p-value: < 2.2e-16Interpreting the results
(Intercept) flipper_length_mm speciesChinstrap speciesGentoo
-4031.4769 40.7054 -206.5101 266.8096 - Intercept: Expected body mass for Adelie penguin with 0 mm flipper length (not meaningful)
- flipper_length_mm: For each 1 mm increase in flipper length, body mass increases by about 40.7 g, holding species constant
- speciesChinstrap: Chinstrap penguins are about 206.5 g lighter than Adelie, holding flipper length constant
- speciesGentoo: Gentoo penguins are about 266.8 g heavier than Adelie, holding flipper length constant
Model diagnostics
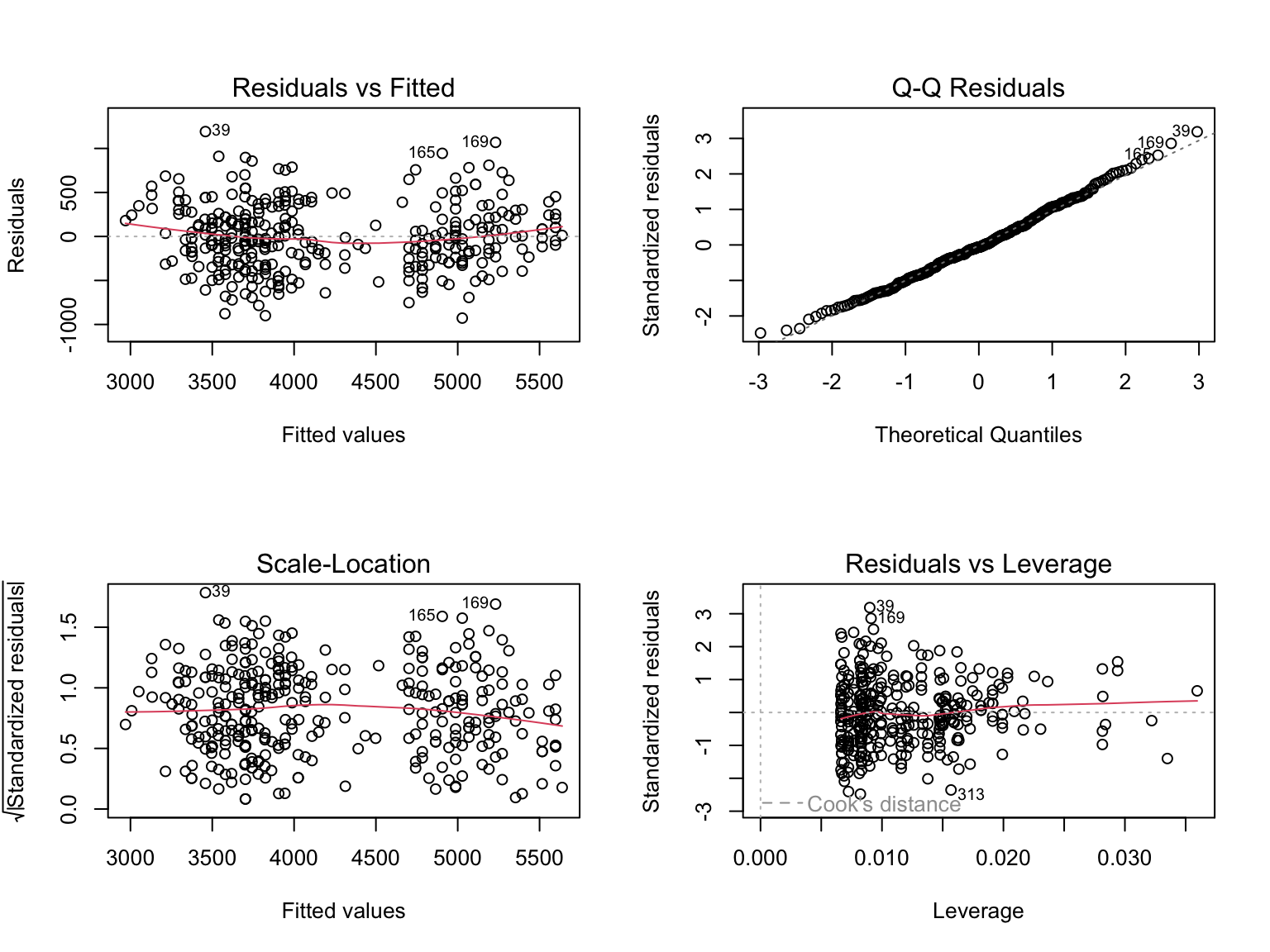
Making predictions
fit lwr upr
1 4783.467 4040.52 5526.413- We predict a Gentoo penguin with 210 mm flippers will have a body mass around 4783 g
- The 95% prediction interval is (4041, 5526) g
The two cultures
Two approaches to statistical modeling
Leo Breiman (2001) identified two cultures in statistical modeling:
Culture 1: Data Modeling
- Focus on inference
- Assume a stochastic model (e.g., linear regression)
- Estimate parameters and test hypotheses
- Interpretability is key
- Assumptions matter greatly
- This lecture has focused here
Culture 2: Algorithmic Modeling
- Focus on prediction
- Treat the data mechanism as unknown
- Prioritize predictive accuracy
- “Black box” models acceptable
- Less concern about distributional assumptions
- The box-office prediction case study has some of this flavor
Implications for your work
- In prediction tasks:
- Diagnostic plots are less critical if the model predicts well on new data
- Large sample sizes can overcome some assumption violations
- Cross-validation and test set performance matter more than p-values
- The “true” model may be unknowable and that’s okay
- In inference tasks (like much of today):
- Understanding relationships is the goal
- Assumptions must be checked carefully
- Coefficient interpretation requires proper model specification
- Causal claims require even stronger assumptions
Summary
Key takeaways
- Regression models relationships between variables
- Simple linear regression: one predictor, estimates slope and intercept
- Multiple regression: multiple predictors, each coefficient represents effect holding others constant
- R function:
lm(response ~ predictors, data = dataset) - Use
summary(),coef(),confint(), andpredict()to interpret results - Check model assumptions using diagnostic plots
- Categorical predictors are handled automatically via dummy variables
- Remember: correlation ≠ causation